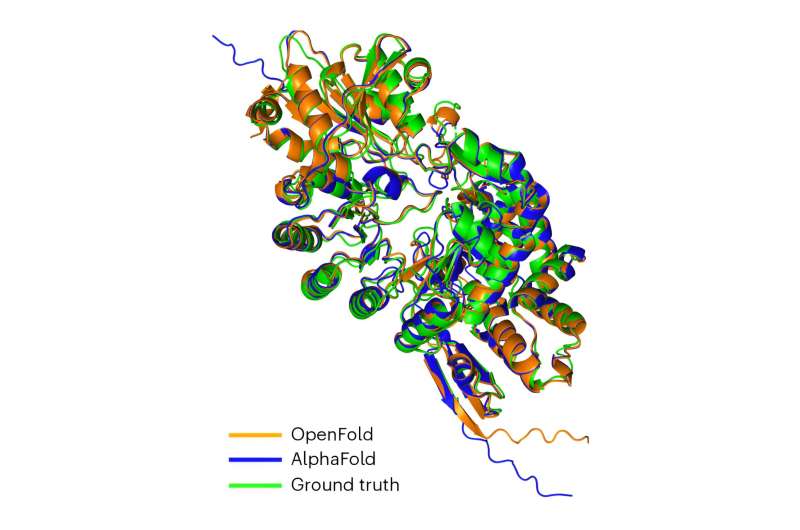
Scientists have developed a new open-source software tool called OpenFold that uses artificial intelligence and harnesses the power of supercomputers to predict protein structures. The image shows that OpenFold matches the accuracy of AlphaFold2 by overlaying predictions from OpenFold and AlphaFold2 with an experimental structure of the Streptomyces tokunonesis TokK protein. Image credit: Natural methods (2024). DOI: 10.1038/s41592-024-02272-z
Form follows function, and this is especially true for the building blocks of life – proteins. The folding and shape of molecular proteins reveal their function in sustaining life.
Scientists have developed a new open-source software tool called OpenFold that uses artificial intelligence (AI) and harnesses the power of supercomputers to predict protein structures.
The research could help develop new drugs and better understand misshapen proteins, such as those associated with neurodegenerative diseases such as Parkinson’s and Alzheimer’s.
OpenFold builds on the success of AlphaFold2, developed by Google DeepMind and used by over two million researchers as of 2021 for protein predictions in vaccine development, cancer treatments, and more.
“AlphaFold2 was a breakthrough for science,” said Nazim Bouatta, a senior research fellow at Harvard Medical School who works at the interface between AI and biology. “We developed a completely open source version – OpenFold – that is now helping academia and industry advance the field.”
Bouatta is co-author of a study in the journal Natural methods Announcing OpenFold, a fast, memory-efficient and trainable implementation of AlphaFold2.
He started the project with his colleague Mohammed AlQuraishi, who used to work at Harvard but now works at Columbia University. The project evolved into the OpenFold Consortium, a consortium of startup companies collaborating with academia.
“Extremely talented students from Harvard and Columbia also contributed to the work, with Gustaf Ahdritz doing remarkable work. They all did a great job implementing the code,” Bouatta said.
A central aspect of AI is the large language models (LLMs) that process huge amounts of text and generate new, meaningful texts from them. One example of this is ChatGPT’s human-like ability to answer queries based on large amounts of text data.
“We need about 100 graphics processing units (GPUs) to train a system like OpenFold. To put it in perspective, to train the latest ChatGPT, you need thousands and thousands of GPUs,” Bouatta said.
One of the very first applications of OpenFold came from Meta AI, formerly Facebook. Meta AI recently published an atlas of more than 600 million proteins from bacteria, viruses, and other microorganisms that had not yet been characterized.
“They used OpenFold to integrate a ‘protein language model’ that is very similar to ChatGPT, but where the language is made up of the amino acids that make up proteins,” Bouatta said.
“In a sense, the information in living organisms is organized in a language,” Bouatta explained, citing the example of the letters ACGT, which represent the four bases of DNA – adenine, cytosine, guanine and thymine. “This is the language that nature has chosen to create these highly evolved living organisms.”
In addition, there is a second level of language for proteins: the letters that represent the 20 amino acids that make up all proteins in the human body and that characterize what function the protein has.
Genome sequencing has produced vast amounts of data on the letters of life, but until now there was no “dictionary” that could calculate the three-dimensional shape of a protein from these letters and model the sites to which small molecules can bind.
“Machine learning allows us to take a string of letters and the amino acids that describe any type of protein imaginable, run a sophisticated algorithm, and return an exquisite three-dimensional structure that is close to what we get from experiments. The OpenFold algorithm is very sophisticated and uses new developments that we see from ChatGPT and others,” Bouatta said, referring to the concepts developed by Google Transformers and elements of ChatGPT’s main algorithm.
A key advantage of OpenFold is the ability to train the model using the scientist’s own data, which is not possible with the publicly available version of AlphaFold2. “The ability to train a system using OpenFold opens up great opportunities for research in both academia and industry,” said Bouatta.
Bouatta expects to release an OpenFold modality in the coming months that can characterize a protein-ligand complex, the intricate alignment of small molecules that bind to a protein.
“This is how medicines work. Understanding this is particularly important,” he explained.
TACC has allocated allocations to the OpenFold team on the Frontera and Lonestar6 supercomputers, particularly the GPU nodes that are critical for developing AI applications worldwide.
“TACC has been an extremely good collaborator,” said Bouatta. “I want to thank TACC for giving us access to these resources so we could use machine learning and AI at the scale we needed.”
“Supercomputers combined with AI are radically changing the way we approach biology. The power of a supercomputer is that it allows us to predict 100 million structures in just a few months. Once the system is trained, we can get structures in seconds. But they will not replace experiments because we need to go back to the lab to test our ideas.”
Integrating AI systems like OpenFold with more traditional, physics-based systems is helping scientists understand life at the most fundamental level and opening up new avenues for treating neurodegenerative diseases.
“Supercomputers are the microscope of the modern era for biology and drug discovery,” Bouatta concluded. “If we continue to put more resources into using the AI/computational approach with supercomputers, we can increase our ability to understand life and cure disease.”
Further information:
Gustaf Ahdritz et al, OpenFold: Retraining AlphaFold2 provides new insights into its learning mechanisms and generalization ability, Natural methods (2024). DOI: 10.1038/s41592-024-02272-z
Provided by the University of Texas at Austin
Quote: AI, computation, and the wrinkles of life: Supercomputers help train a software tool for the protein modeling community (August 13, 2024), accessed August 13, 2024, from https://phys.org/news/2024-08-ai-life-supercomputers-software-tool.html
This document is subject to copyright. Except for the purposes of private study or research, no part of it may be reproduced without written permission. The contents are for information purposes only.

